The DC method is a robust scheme and can be applicable to a wide variety
of materials with a reasonable degree of accuracy and efficiency, while
this scheme is suitable especially for covalent systems.
In this subsection, the O(![]() ) calculation using the DC method is
illustrated. In an input file 'DIA64_DC.dat' which can be found in the
directory 'work', please specify DC for
the keyword 'scf.EigenvalueSolver'.
) calculation using the DC method is
illustrated. In an input file 'DIA64_DC.dat' which can be found in the
directory 'work', please specify DC for
the keyword 'scf.EigenvalueSolver'.
scf.EigenvalueSolver DC
% ./openmx DIA64_DC.dat
The accuracy and efficiency of the DC method are controlled by two simple parameters: 'orderN.HoppingRanges' and 'orderN.NumHoppings'.
|
The keyword 'orderN.HoppingRanges' defines the radius of a sphere which is centered on each atom. The logically truncated cluster for each atom is constructed for the atoms inside the sphere.
The keyword 'orderN.NumHoppings' gives
the number, ![]() , of hopping
which is required to construct the logically truncated cluster.
The cluster of size,
, of hopping
which is required to construct the logically truncated cluster.
The cluster of size, ![]() , is defined by all neighbors that can be reached
by
, is defined by all neighbors that can be reached
by ![]() hops, where the cutoff distance is given by the sum of the cutoff
distances
hops, where the cutoff distance is given by the sum of the cutoff
distances ![]() and
and ![]() of basis orbitals belonging to atoms 1 and 2.
of basis orbitals belonging to atoms 1 and 2.
If the number of atoms in the systems is N, N small eigenvalue problems for the N logically truncated clusters are solved, and then the total density of states (DOS) is constructed as the sum of the projected DOS of each logically truncated cluster. Although the appropriate values for 'orderN.HoppingRanges' and 'orderN.NumHoppings' depend on interested systems, for molecular systems the following values are recommended as a trade-off between the computational accuracy and efficiency:
orderN.HoppingRanges 6.0 - 7.0
orderN.NumHoppings 2
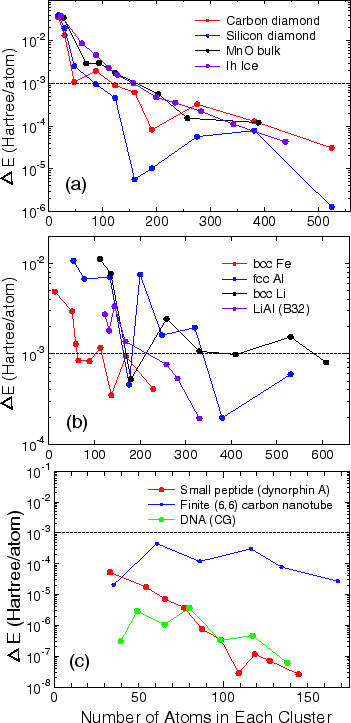 |
Table 2 shows the comparison in the total energy between the exact
diagonalization and the DC method for a C![]() molecule and small
peptide molecules (valorphin [52]), and DNA consisting of
cytosines and guanines.
We find that errors in the total energy calculated by the DC method are
about a few mHartree in this system size.
Also, it can be estimated that the DC method is faster
than the conventional diagonalization when the number of atoms is
larger than 500 atoms, while the crossing point between the conventional
diagonalization and the DC method with respect to computational time
depends on systems and the number of processors in parallel implementation.
molecule and small
peptide molecules (valorphin [52]), and DNA consisting of
cytosines and guanines.
We find that errors in the total energy calculated by the DC method are
about a few mHartree in this system size.
Also, it can be estimated that the DC method is faster
than the conventional diagonalization when the number of atoms is
larger than 500 atoms, while the crossing point between the conventional
diagonalization and the DC method with respect to computational time
depends on systems and the number of processors in parallel implementation.
To see an overall tendency in the convergence properties of total energy with respect to the size of truncated cluster, the error in the total energy, compared to the exact diagonalization, is shown as a function of the number of atoms in each cluster for (a) bulks with a finite gap, (b) metals, and (c) molecular systems in Fig. 16. We see that the error decreases almost exponentially for the bulks with a finite gap and molecular systems, while the convergence speed is slower for metals.