 |
The basis orbitals can be variationally optimized using the orbital optimization method [23]. As an illustration of the orbital optimization, let us explain using a methane molecule of which input file is Methane_OO.dat. The following keywords in this file are set as follows:
<Definition.of.Atomic.Species
H H4.0-s41p41 H_TM
C C4.5-s41p41 C_TM_PCC
Definition.of.Atomic.Species>
orbitalOpt.Method species # Off|Unrestricted|Restricted
orbitalOpt.InitCoes Symmetrical # Symmetrical|Free
orbitalOpt.initPrefactor 0.1 # default=0.1
orbitalOpt.scf.maxIter 25 # default=12
orbitalOpt.MD.maxIter 10 # default=5
orbitalOpt.per.MDIter 20 # default=1000000
orbitalOpt.criterion 1.0e-6 # default=1.0e-4 (Hartree/borh)^2
Num.CntOrb.Atoms 2 # default=1
<Atoms.Cont.Orbitals
1
2
Atoms.Cont.Orbitals>
Then, we execute OpenMX as:
% ./openmx Methane.dat
When the execution is completed normally, you can find the history
of orbital optimization in the file 'met_oo.out' as:
***********************************************************
***********************************************************
History of orbital optimization MD= 1
********* Gradient Norm ((Hartree/borh)^2) ********
Required criterion= 0.000001000000
***********************************************************
iter= 1 Gradient Norm= 0.081251614657 Uele= -2.750500719281
iter= 2 Gradient Norm= 0.018543400953 Uele= -2.933260690003
iter= 3 Gradient Norm= 0.005918002913 Uele= -2.966113950591
iter= 4 Gradient Norm= 0.001553729359 Uele= -3.010077558163
iter= 5 Gradient Norm= 0.000356946294 Uele= -3.012729963043
iter= 6 Gradient Norm= 0.000119196944 Uele= -3.024577717351
iter= 7 Gradient Norm= 0.000042934968 Uele= -3.024772396249
iter= 8 Gradient Norm= 0.000031243105 Uele= -3.026624698820
iter= 9 Gradient Norm= 0.000020515771 Uele= -3.026569330230
iter= 10 Gradient Norm= 0.000015126154 Uele= -3.026833093004
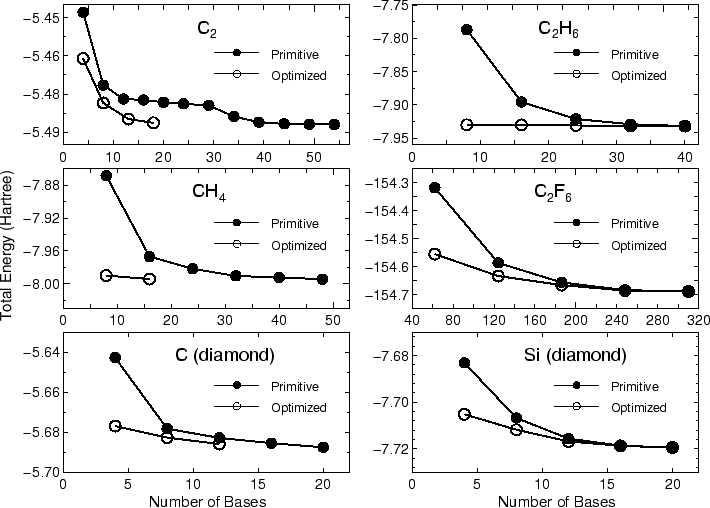
In most cases, ten iterative steps are enough to achieve
a sufficient convergence. The comparison between the primitive basis
orbitals and the optimized orbitals in the total energy
is given by
Primitive basis orbitals
Utot = -8.032594073571 (Hartree)
Optimized orbitals by the orbital optimization
Utot = -8.150139929748 (Hartree)
 |
The following three options are available for the keyword 'orbitalOpt.Method', the unrestricted optimization 'Unrestricted', the restricted optimization 'Restricted', and Orbital optimization restricted to species 'Species'.
The radial functions of basis orbitals are optimized without any constraint. Thus, all the radial functions could differ from each other, which could depend on the following indices, atomic number, angular moment quantum number, magnetic quantum number, and orbital multiplicity.
The radial functions of basis orbitals are optimized
with a constraint that the radial wave function ![]() is independent
on the magnetic quantum number. We prefer 'Restricted' to 'Unrestricted',
since the restricted optimization guarantees the rotational invariance
of the total energy.
is independent
on the magnetic quantum number. We prefer 'Restricted' to 'Unrestricted',
since the restricted optimization guarantees the rotational invariance
of the total energy.
Basis orbitals in atoms with the same species name, that you define in 'Definition.of.Atomic.Species', are optimized as the same orbitals. If you want to assign the same orbitals to atoms with almost the same chemical environment, and optimize these orbitals, this scheme could be quite convenient.