 |
The radial function of basis orbitals can be variationally optimized using the orbital optimization method [26]. As an illustration of the orbital optimization, let us explain it using a methane molecule of which input file is Methane_OO.dat. In the orbital optimization method the optimized orbitals are expressed by the linear combination of primitive orbitals, and obtained by variationally optimizing the contraction coefficients. The number of the primitive and optimized orbitals in the optimization are specified by
<Definition.of.Atomic.Species
H H5.0-s4>1 H_CA11
C C5.0-s4>1p4>1 C_CA11
Definition.of.Atomic.Species>
orbitalOpt.Method species # Off|Species|Atoms
orbitalOpt.Opt.Method EF # DIIS|EF
orbitalOpt.SD.step 0.001 # default=0.001
orbitalOpt.HistoryPulay 30 # default=15
orbitalOpt.StartPulay 10 # default=1
orbitalOpt.scf.maxIter 60 # default=40
orbitalOpt.Opt.maxIter 140 # default=100
orbitalOpt.per.MDIter 20 # default=1000000
orbitalOpt.criterion 1.0e-4 # default=1.0e-4
CntOrb.fileout on # on|off, default=off
Num.CntOrb.Atoms 2 # default=1
<Atoms.Cont.Orbitals
1
2
Atoms.Cont.Orbitals>
% ./openmx Methane_OO.dat
***********************************************************
***********************************************************
History of orbital optimization MD= 1
********* Gradient Norm ((Hartree/borh)^2) ********
Required criterion= 0.000100000000
***********************************************************
iter= 1 Gradient Norm= 0.057099116186 Uele= -3.217165688850
iter= 2 Gradient Norm= 0.044668590658 Uele= -3.220124728727
iter= 3 Gradient Norm= 0.034308424834 Uele= -3.223127810870
iter= 4 Gradient Norm= 0.025847684906 Uele= -3.226182452559
iter= 5 Gradient Norm= 0.019106482737 Uele= -3.229299561866
iter= 6 Gradient Norm= 0.013893901444 Uele= -3.232493788494
iter= 7 Gradient Norm= 0.010499563363 Uele= -3.235308811420
iter= 8 Gradient Norm= 0.008362684115 Uele= -3.237657609355
iter= 9 Gradient Norm= 0.006959746319 Uele= -3.239623291338
iter= 10 Gradient Norm= 0.005994856595 Uele= -3.241273216875
iter= 11 Gradient Norm= 0.005298132389 Uele= -3.242661771498
iter= 12 Gradient Norm= 0.003059669828 Uele= -3.250897736701
iter= 13 Gradient Norm= 0.001390212138 Uele= -3.255127796347
iter= 14 Gradient Norm= 0.000781057925 Uele= -3.255185826155
iter= 15 Gradient Norm= 0.000726780237 Uele= -3.255269791193
iter= 16 Gradient Norm= 0.000390997436 Uele= -3.250876275966
iter= 17 Gradient Norm= 0.000280772241 Uele= -3.250340948913
iter= 18 Gradient Norm= 0.000200449371 Uele= -3.252358609122
iter= 19 Gradient Norm= 0.000240967733 Uele= -3.254237590017
iter= 20 Gradient Norm= 0.000081978704 Uele= -3.258145769943
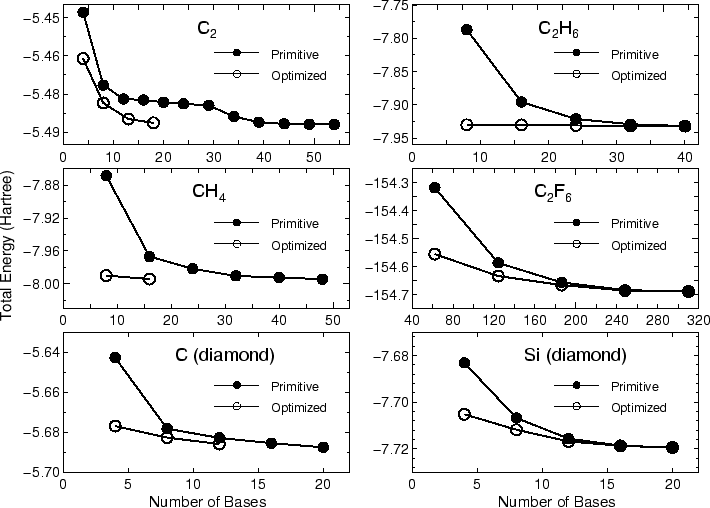
In most cases, 20-50 iterative steps are enough to achieve
a sufficient convergence. The comparison between the primitive basis
orbitals and the optimized orbitals in the total energy
is given by
Primitive basis orbitals
Utot = -7.992568903114 (Hartree)
Optimized orbitals by the orbital optimization
Utot = -8.133746206187 (Hartree)
 |
The following two options are available for the keyword 'orbitalOpt.Method', 'atoms' in which basis obitals on each atom are fully optimized, 'species' in which basis obitals on each species are optimized.
The radial functions of basis orbitals are optimized
with a constraint that the radial wave function ![]() is independent
on the magnetic quantum number, which guarantees the rotational invariance
of the total energy. However, the optimized orbital on all the atoms can
be different eath other.
is independent
on the magnetic quantum number, which guarantees the rotational invariance
of the total energy. However, the optimized orbital on all the atoms can
be different eath other.
Basis orbitals in atoms with the same species name, that you define in 'Definition.of.Atomic.Species', are optimized as the same orbitals. If you want to assign the same orbitals to atoms with almost the same chemical environment, and optimize these orbitals, this scheme could be quite convenient.
Although the same information is available in the section 'Input file',
for convenience the details of the other keywords are listed below:
orbitalOpt.scf.maxIter
The maximum number of SCF iterations in the orbital optimization is
specified by the keyword 'orbitalOpt.scf.maxIter'.
orbitalOpt.Opt.maxIter
The maximum number of iterations for the orbital optimization is specified
by the keyword 'orbitalOpt.Opt.maxIter'. The iteration loop for the orbital
optimization is terminated at the number specified by 'orbitalOpt.Opt.maxIter'
even when a convergence criterion is not satisfied.
orbitalOpt.Opt.Method
Two schemes for the optimization of orbitals are available:
'EF' which is an eigenvector following method, 'DIIS' which is
the direct inversion method in iterative subspace.
The algorithms are basically same as for the geometry optimization.
Either 'EF' or 'DIIS' is chosen by the keyword, 'orbitalOpt.Opt.Method'.
orbitalOpt.StartPulay
The quasi Newton method, 'EF' and 'DIIS' starts from the optimization step
specified by the keyword 'orbitalOpt.StartPulay'.
orbitalOpt.HistoryPulay
The keyword 'orbitalOpt.HistoryPulay' specifies the number of previous steps
to estimate the next input contraction coefficients used in the quasi Newton
method, 'EF' and 'DIIS'.
orbitalOpt.SD.step
Steps before moving the quasi Newton method, 'EF' and 'DIIS' is performed
by the steepest decent method. The prefactor used in the steepest decent
method is specified by the keyword 'orbitalOpt.SD.step'. In most cases,
orbitalOpt.SD.step of 0.001 can be a good prefactor.
orbitalOpt.criterion
The keyword 'orbitalOpt.criterion' specifies a convergence criterion
((Hartree/borh)![]() ) for the orbital optimization. The iterations loop is
finished when a condition, Norm of derivatives
) for the orbital optimization. The iterations loop is
finished when a condition, Norm of derivatives![]() orbitalOpt.criterion,
is satisfied.
orbitalOpt.criterion,
is satisfied.
CntOrb.fileout
If you want to output the optimized radial orbitals to files,
then the keyword 'CntOrb.fileout' must be ON.
Num.CntOrb.Atoms
The keyword 'Num.CntOrb.Atoms' gives the number of atoms whose
optimized radial orbitals are output to files.
Atoms.Cont.Orbitals
The keyword 'Atoms.Cont.Orbitals' specifies the atom number,
which was given by the first column in the specification of
the keyword 'Atoms.SpeciesAndCoordinates' for the output
of optimized orbitals as follows:
<Atoms.Cont.Orbitals
1
2
Atoms.Cont.Orbitals>
The beginning of the description must be '